With the recent success of mRNA therapeutics including the mRNA vaccines used during the pandemic, we have a therapeutic tool that can adjust gene expression or alternatively can be used for vaccines presenting antigens to both the cell-mediated and humoral immune response. My research programme is dedicated to the development of computational tools used to design these therapies and vaccines in cancer and other diseases.
Our scientific understanding of the human immune-system is now progressing at a rapid rate, allowing for the development of computational models that could predict the immune-systems of specific individuals and populations. Simultaneously, the last 2 months have seen breakthroughs in our ability to leverage Artificial Intelligence and Natural Language Processing techniques to design proteins with new functions, with tremendous implications for synthetic protein drug design and vaccine design. I am particularly interested in how these fields will converge through mRNA therapeutics and vaccine technologies. In order for these therapeutics to work, the designed proteins must avoid immune clearance or other immune-related toxicities. And for vaccines, the opposite is true, the immune-response must be broad across a defined population. In addition, mRNA therapeutics must conform to design constraints provided by manufacturers and ofcourse must express well in order to be functional. For these reasons there are interesting computational challenges in designing mRNA therapeutics and vaccines that require integrative -omics to identify drug targets, to develop theranostic and prognostic biomarkers for precision medicine, to survey different facets of immunity and keep progressing predictive immunology, and to combine this knowledge with AI-methods computational protein and mRNA design to progress the field of mRNA therapeutic or vaccine design.
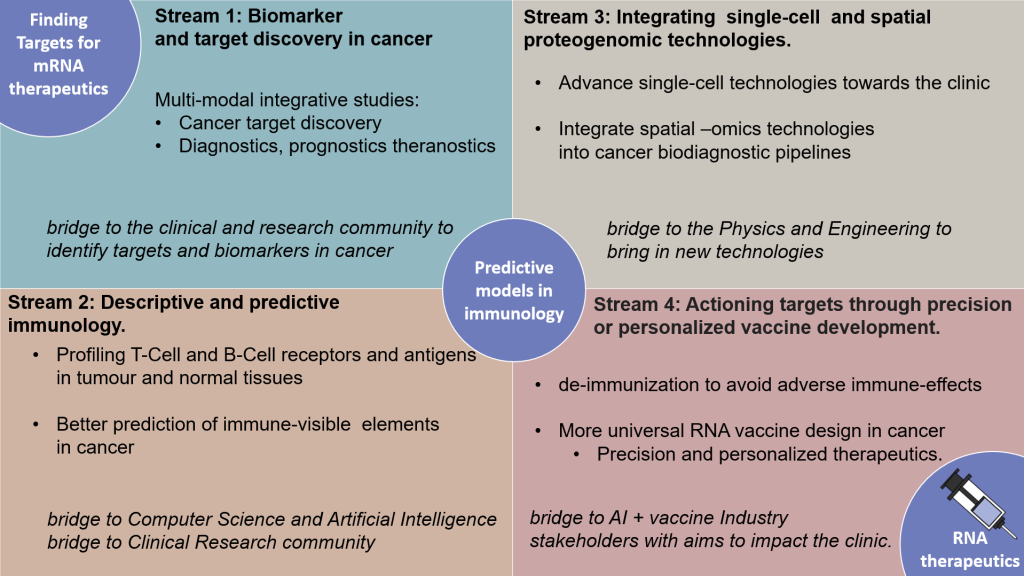
In short, I am developing a highly collaborative and translational research programme dedicated to the identification of prognostic/theranostic biomarkers and therapeutic targets in complex diseases including cancers towards the development of new lines of RNA therapeutics to treat these diseases. My research leverages multi-omic interrogation of mass-spectrometry and high-throughput sequencing data and is progressing towards single cells. Because better predictive immunology models are fundamental to mRNA therapeutics and vaccines, high-throughput methods to interrogate the immune-disease synapse will be fundamental to the programme. The combination of the different streams described here will lead to the eventual development of AI-driven tools for mRNA therapeutic design, where the goal is to produce end-to-end solutions to deliver new protein drugs and vaccines as mRNA constructs with defined immunological profiles. I outline four interrelated research streams below, with a few publications.